In this post I’ll be taking a break from my normal explorations in the medical device domain to talk about Sliced. Sliced is a 2-hour data science competition streamed on Twitch and hosted by Meg Risdal and Nick Wan. Four competitors tackle a prediction problem in real time using whatever coding language or tools they prefer, grabbing bonus points along the way for Data Visualization and/or stumbling onto Golden Features (hint: always calculate the air density when training on weather data). Viewers can simply kick back on watch the contestants apply their trade or they can actively participate by submitting their own solutions and seeing how they stack up on the competition leaderboard!
Here are my observations after watching a few episodes:
* Participants do not typically implement more than 2 different model types, preferring to spend their time on Feature Engineering and tuning the hyperparameters of their preferred model
* Gradient boosting (XGBoost, Catboost, etc) is the dominant technique for tabular data
To clarify the first point above - the tuning is not totally manual; grid search functions are typically employed to identify the best hyperparameters from a superset of options. But the time pressure of the competition means that players can’t set up massive grids that lock up compute resources for too long. So it’s generally an iterative process over small grids that are expanded and contracted as needed based on intermediate results of the model predicting on a test set.
All this led me to wonder: given the somewhat manual process of hyperparameter optimization and the restricted number of model types… how would AutoML fare in Sliced? The rest of this post will attempt to answer that question, at least for an arbitrary Sliced episode.
- Setup
- Understanding the Ensemble
- Final Thoughts
- Session Info
- Appendix - Full Details of the Ensemble Model
Setup
For this exercise we’ll use the dataset and metrics from Episode 7 in which we are asked to predict whether or not a bank customer churned. The scoring metric is LogLoss. I’ll be using the free version of the h2o.ai framework and take the following approach to feature engineering and variable selection:
- All variables will be used (churn explained by everything) and no feature engineering except imputing means for missing values, converting nominal predictors to dummy variables, and removing ID column. This should give a fair look at how h2o will do given the bare minimum of attention to pre-processing and no attention to model selection or hyperparameter range.
Let’s get to it.
Load libraries
library(tidymodels)
library(tidyverse)
library(h2o)
library(here)
library(gt)Load dataset
Read the data in as a csv and rename the attrition column
dataset <- read_csv(here("ep7/train.csv")) %>%
mutate(churned = case_when(
attrition_flag == 1 ~ "yes",
TRUE ~ "no"
) %>% as_factor()) %>%
select(-attrition_flag)
holdout <- read_csv(here("ep7/test.csv"))
dataset %>%
gt_preview() %>%
cols_align("center")| id | customer_age | gender | education_level | income_category | total_relationship_count | months_inactive_12_mon | credit_limit | total_revolving_bal | total_amt_chng_q4_q1 | total_trans_amt | total_trans_ct | total_ct_chng_q4_q1 | avg_utilization_ratio | churned | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 8805 | 27 | F | Post-Graduate | Less than $40K | 3 | 2 | 1438.3 | 990 | 0.715 | 3855 | 73 | 1.147 | 0.688 | no |
| 2 | 4231 | 42 | F | College | Less than $40K | 6 | 4 | 3050.0 | 1824 | 0.771 | 1973 | 50 | 1.381 | 0.598 | no |
| 3 | 5263 | 47 | F | Unknown | Less than $40K | 3 | 3 | 1561.0 | 0 | 0.502 | 1947 | 28 | 0.556 | 0.000 | yes |
| 4 | 2072 | 44 | M | Uneducated | $80K - $120K | 1 | 3 | 25428.0 | 1528 | 0.725 | 13360 | 97 | 0.796 | 0.060 | no |
| 5 | 7412 | 54 | M | Graduate | $60K - $80K | 3 | 3 | 2947.0 | 2216 | 0.760 | 1744 | 53 | 0.606 | 0.752 | no |
| 6..7087 | |||||||||||||||
| 7088 | 7932 | 57 | F | High School | Less than $40K | 5 | 3 | 3191.0 | 2517 | 0.719 | 1501 | 35 | 0.591 | 0.789 | no |
Set Up Basic Recipe
This recipe starts with the model structure, imputes the mean for numeric predictors, and converts nominal variables to dummy. Id is also removed. Note that churned is described by . which means “all variables”.
basic_rec <- recipe(churned ~ .,
data = dataset
) %>%
step_impute_mean(all_numeric_predictors()) %>%
step_dummy(all_nominal_predictors()) %>%
step_rm(id)Prep and Bake the Dataset
Prepping and baking functions will apply the recipe to the supplied dataset and make it usable in tibble format for passing to h2o.
baked_dataset_tbl <- basic_rec %>%
prep() %>%
bake(dataset)
baked_dataset_tbl %>%
gt_preview() %>%
cols_align("center")| customer_age | total_relationship_count | months_inactive_12_mon | credit_limit | total_revolving_bal | total_amt_chng_q4_q1 | total_trans_amt | total_trans_ct | total_ct_chng_q4_q1 | avg_utilization_ratio | churned | gender_M | education_level_Doctorate | education_level_Graduate | education_level_High.School | education_level_Post.Graduate | education_level_Uneducated | education_level_Unknown | income_category_X.40K....60K | income_category_X.60K....80K | income_category_X.80K....120K | income_category_Less.than..40K | income_category_Unknown | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 27 | 3 | 2 | 1438.3 | 990 | 0.715 | 3855 | 73 | 1.147 | 0.688 | no | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 2 | 42 | 6 | 4 | 3050.0 | 1824 | 0.771 | 1973 | 50 | 1.381 | 0.598 | no | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 3 | 47 | 3 | 3 | 1561.0 | 0 | 0.502 | 1947 | 28 | 0.556 | 0.000 | yes | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| 4 | 44 | 1 | 3 | 25428.0 | 1528 | 0.725 | 13360 | 97 | 0.796 | 0.060 | no | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| 5 | 54 | 3 | 3 | 2947.0 | 2216 | 0.760 | 1744 | 53 | 0.606 | 0.752 | no | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 6..7087 | |||||||||||||||||||||||
| 7088 | 57 | 5 | 3 | 3191.0 | 2517 | 0.719 | 1501 | 35 | 0.591 | 0.789 | no | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
Working in h2o
Convert Dataset to h2o
h2o must first be initialized and then the data can be coerced to h2o type. Using the h2o.describe() function shows a nice summary of the dataset and verifies that it was imported correctly.
h2o.init()## Connection successful!
##
## R is connected to the H2O cluster:
## H2O cluster uptime: 8 hours 42 minutes
## H2O cluster timezone: America/Los_Angeles
## H2O data parsing timezone: UTC
## H2O cluster version: 3.32.1.3
## H2O cluster version age: 2 months and 5 days
## H2O cluster name: H2O_started_from_R_kingr17_afp345
## H2O cluster total nodes: 1
## H2O cluster total memory: 0.85 GB
## H2O cluster total cores: 4
## H2O cluster allowed cores: 4
## H2O cluster healthy: TRUE
## H2O Connection ip: localhost
## H2O Connection port: 54321
## H2O Connection proxy: NA
## H2O Internal Security: FALSE
## H2O API Extensions: Amazon S3, Algos, AutoML, Core V3, TargetEncoder, Core V4
## R Version: R version 4.0.3 (2020-10-10)train_h2_tbl <- as.h2o(baked_dataset_tbl)##
|
| | 0%
|
|======================================================================| 100%h2o.describe(train_h2_tbl) %>%
gt() %>%
cols_align("center")| Label | Type | Missing | Zeros | PosInf | NegInf | Min | Max | Mean | Sigma | Cardinality |
|---|---|---|---|---|---|---|---|---|---|---|
| customer_age | int | 0 | 0 | 0 | 0 | 26.0 | 73.000 | 4.630742e+01 | 7.9929340 | NA |
| total_relationship_count | int | 0 | 0 | 0 | 0 | 1.0 | 6.000 | 3.818426e+00 | 1.5516740 | NA |
| months_inactive_12_mon | int | 0 | 22 | 0 | 0 | 0.0 | 6.000 | 2.331546e+00 | 1.0103784 | NA |
| credit_limit | real | 0 | 0 | 0 | 0 | 1438.3 | 34516.000 | 8.703875e+03 | 9152.3072697 | NA |
| total_revolving_bal | int | 0 | 1705 | 0 | 0 | 0.0 | 2517.000 | 1.169674e+03 | 815.4734902 | NA |
| total_amt_chng_q4_q1 | real | 0 | 5 | 0 | 0 | 0.0 | 3.355 | 7.601713e-01 | 0.2213876 | NA |
| total_trans_amt | int | 0 | 0 | 0 | 0 | 563.0 | 18484.000 | 4.360548e+03 | 3339.1008390 | NA |
| total_trans_ct | int | 0 | 0 | 0 | 0 | 10.0 | 139.000 | 6.465378e+01 | 23.3431055 | NA |
| total_ct_chng_q4_q1 | real | 0 | 7 | 0 | 0 | 0.0 | 3.500 | 7.126414e-01 | 0.2363882 | NA |
| avg_utilization_ratio | real | 0 | 1705 | 0 | 0 | 0.0 | 0.999 | 2.753678e-01 | 0.2755652 | NA |
| churned | enum | 0 | 5956 | 0 | 0 | 0.0 | 1.000 | 1.597065e-01 | 0.3663595 | 2 |
| gender_M | int | 0 | 3714 | 0 | 0 | 0.0 | 1.000 | 4.760158e-01 | 0.4994597 | NA |
| education_level_Doctorate | int | 0 | 6762 | 0 | 0 | 0.0 | 1.000 | 4.599323e-02 | 0.2094852 | NA |
| education_level_Graduate | int | 0 | 4876 | 0 | 0 | 0.0 | 1.000 | 3.120767e-01 | 0.4633737 | NA |
| education_level_High.School | int | 0 | 5710 | 0 | 0 | 0.0 | 1.000 | 1.944131e-01 | 0.3957761 | NA |
| education_level_Post.Graduate | int | 0 | 6745 | 0 | 0 | 0.0 | 1.000 | 4.839165e-02 | 0.2146075 | NA |
| education_level_Uneducated | int | 0 | 6050 | 0 | 0 | 0.0 | 1.000 | 1.464447e-01 | 0.3535764 | NA |
| education_level_Unknown | int | 0 | 6005 | 0 | 0 | 0.0 | 1.000 | 1.527935e-01 | 0.3598137 | NA |
| income_category_X.40K....60K | int | 0 | 5865 | 0 | 0 | 0.0 | 1.000 | 1.725451e-01 | 0.3778802 | NA |
| income_category_X.60K....80K | int | 0 | 6064 | 0 | 0 | 0.0 | 1.000 | 1.444695e-01 | 0.3515900 | NA |
| income_category_X.80K....120K | int | 0 | 6007 | 0 | 0 | 0.0 | 1.000 | 1.525113e-01 | 0.3595411 | NA |
| income_category_Less.than..40K | int | 0 | 4621 | 0 | 0 | 0.0 | 1.000 | 3.480530e-01 | 0.4763865 | NA |
| income_category_Unknown | int | 0 | 6316 | 0 | 0 | 0.0 | 1.000 | 1.089165e-01 | 0.3115564 | NA |
Specify the Response and Predictors
In h2o we must identify the response column and the predictors which we do here. Unfortunately we can’t tidyselect here I don’t think.
y <- "churned"
x <- setdiff(names(train_h2_tbl), y)autoML Search and Optimization
Now we start the autoML session. You can specify the stopping rule by either total number of model or total duration in seconds. Since we’re simulating a timed Sliced competition, we’ll use max time. The longest I observed competitors training for was about 20 minutes, so we’ll use that here and then grab a coffee while it chugs. Notice that in this API we are not specifying any particular type of model or any hyperparameter range to optimize over.
# aml <- h2o.automl(
# y = y,
# x = x,
# training_frame = train_h2_tbl,
# project_name = "sliced_ep7_refactored_25bjuly2021",
# max_runtime_secs = 1200,
# seed = 07252021
# )Leaderboard of Best Model
Not to be confused with the competition leaderboard, h2o will produce a “leaderboard” of models that it evaluated and ranked as part of its session. Here we access and observe the leaderboard and its best models.
# leaderboard_tbl <- aml@leaderboard %>% as_tibble()
# a <- leaderboard_tbl %>% gt_preview()
# a| model_id | auc | logloss | aucpr | mean_per_class_error | rmse | mse | |
|---|---|---|---|---|---|---|---|
| 1 | StackedEnsemble_AllModels_AutoML_20210725_011103 | 0.9918194 | 0.08285245 | 0.9636402 | 0.06007334 | 0.1543060 | 0.02381035 |
| 2 | GBM_2_AutoML_20210725_011103 | 0.9914487 | 0.08636589 | 0.9626320 | 0.05620012 | 0.1569434 | 0.02463124 |
| 3 | StackedEnsemble_BestOfFamily_AutoML_20210725_011103 | 0.9914280 | 0.08451327 | 0.9625496 | 0.05620012 | 0.1556844 | 0.02423764 |
| 4 | GBM_grid__1_AutoML_20210725_011103_model_2 | 0.9913165 | 0.09009359 | 0.9616467 | 0.06286946 | 0.1600684 | 0.02562190 |
| 5 | GBM_4_AutoML_20210725_011103 | 0.9912436 | 0.09035254 | 0.9602742 | 0.07244469 | 0.1599096 | 0.02557107 |
| 6..37 | |||||||
| 38 | GLM_1_AutoML_20210725_011103 | 0.9152440 | 0.24794654 | 0.7267223 | 0.18132441 | 0.2727786 | 0.07440815 |
Extract Top Model
As expected, the top slot is an ensemble (in my limited experience it usually is). This is the one we’ll use to predict churn on the holdout set and submit to kaggle for our competition results. The ensemble model is extracted and stored as follows:
# model_names <- leaderboard_tbl$model_id
# top_model <- h2o.getModel(model_names[1])To prepare the holdout data predictions, we apply the basic recipe and convert to h2o, just as before with the training data.
Pre-Process the Holdout Set
We want the basic recipe applied to the holdout set so the model sees the same type of predictor variables when it goes to make predictions.
holdout_h2o_tbl <- basic_rec %>%
prep() %>%
bake(holdout) %>%
as.h2o()##
|
| | 0%
|
|======================================================================| 100%Make Predictions
Predictions are made using h2o.predict().
top_model_basic_preds <- h2o.predict(top_model, newdata = holdout_h2o_tbl) %>%
as_tibble() %>%
bind_cols(holdout) %>%
select(id, attrition_flag = yes)##
|
| | 0%
|
|======================================================================| 100%top_model_basic_preds %>%
gt_preview() | id | attrition_flag | |
|---|---|---|
| 1 | 3005 | 0.0020805123 |
| 2 | 143 | 0.8462792238 |
| 3 | 5508 | 0.0003131364 |
| 4 | 6474 | 0.0001977184 |
| 5 | 9784 | 0.0017571210 |
| 6..3038 | ||
| 3039 | 9822 | 0.9895064264 |
Nice! A list of predictions for each id in the holdout set. Let’s write it to file and submit.
Export Results
top_model_basic_preds %>%
write_csv(here("run1_basic_ep7.csv"))Scoring the Submission
To submit the file for assessment, simply upload the csv to the interface on kaggle by browsing and selecting or dragging. In a few seconds our LogLoss on holdout set is revealed. The one we care about is the private score.

A private score of 0.06921! Would it have won the modeling portion of the competition? No. But it appears to be a very efficient way to get reasonably close to the goal. This submission would have slotted me into 3rd place out of 31 entries, just behind eventual Episode 7 winner Ethan Douglas. And remember, I didn’t need to specify any modeling engine or range of hyperparameters to tune!
Understanding the Ensemble
Just so we aren’t totally naive, let’s dig into this model a bit and see what h2o built. For an ensemble, we want to interrogate the “metalearner” which can be thought of as the model that is made up of many models. These are S4 objects which require the @ operator to dig into the different slots / items (I just learned this about 90 seconds ago).
Extract the Ensemble Metalearner Model
metalearner_model <- h2o.getModel(top_model@model$metalearner$name)
metalearner_model@model$model_summary %>%
as_tibble() %>%
gt_preview() %>%
cols_align("center")| family | link | regularization | number_of_predictors_total | number_of_active_predictors | number_of_iterations | training_frame | |
|---|---|---|---|---|---|---|---|
| 1 | binomial | logit | Elastic Net (alpha = 0.5, lambda = 0.00311 ) | 36 | 12 | 7 | levelone_training_StackedEnsemble_AllModels_AutoML_20210725_011103 |
Looks like we have 36 different models combined in a GLM, using Elastic Net regularization. The component models are either GBM or Deep Learning and it looks like they each have different hyperparameter grids that were searched across. I’ll put the full model output in the Appendix. Here are a list of the models that compose the ensemble and their relative importance to the ensemble. The top performing models take a greater weight.
Importance of Each Contributing Model in the Ensemble
h2o.varimp(metalearner_model) %>%
as_tibble() %>%
gt_preview() %>%
cols_align("center")| variable | relative_importance | scaled_importance | percentage | |
|---|---|---|---|---|
| 1 | GBM_2_AutoML_20210725_011103 | 0.7014266 | 1.0000000 | 0.16199734 |
| 2 | GBM_grid__1_AutoML_20210725_011103_model_18 | 0.6565727 | 0.9360534 | 0.15163815 |
| 3 | GBM_grid__1_AutoML_20210725_011103_model_9 | 0.6336205 | 0.9033312 | 0.14633725 |
| 4 | GBM_grid__1_AutoML_20210725_011103_model_19 | 0.4374311 | 0.6236307 | 0.10102651 |
| 5 | GBM_grid__1_AutoML_20210725_011103_model_3 | 0.4047249 | 0.5770026 | 0.09347288 |
| 6..35 | ||||
| 36 | GLM_1_AutoML_20210725_011103 | 0.0000000 | 0.0000000 | 0.00000000 |
We could also visualize the data above for scaled importance of each model within the ensemble:
h2o.varimp_plot(metalearner_model)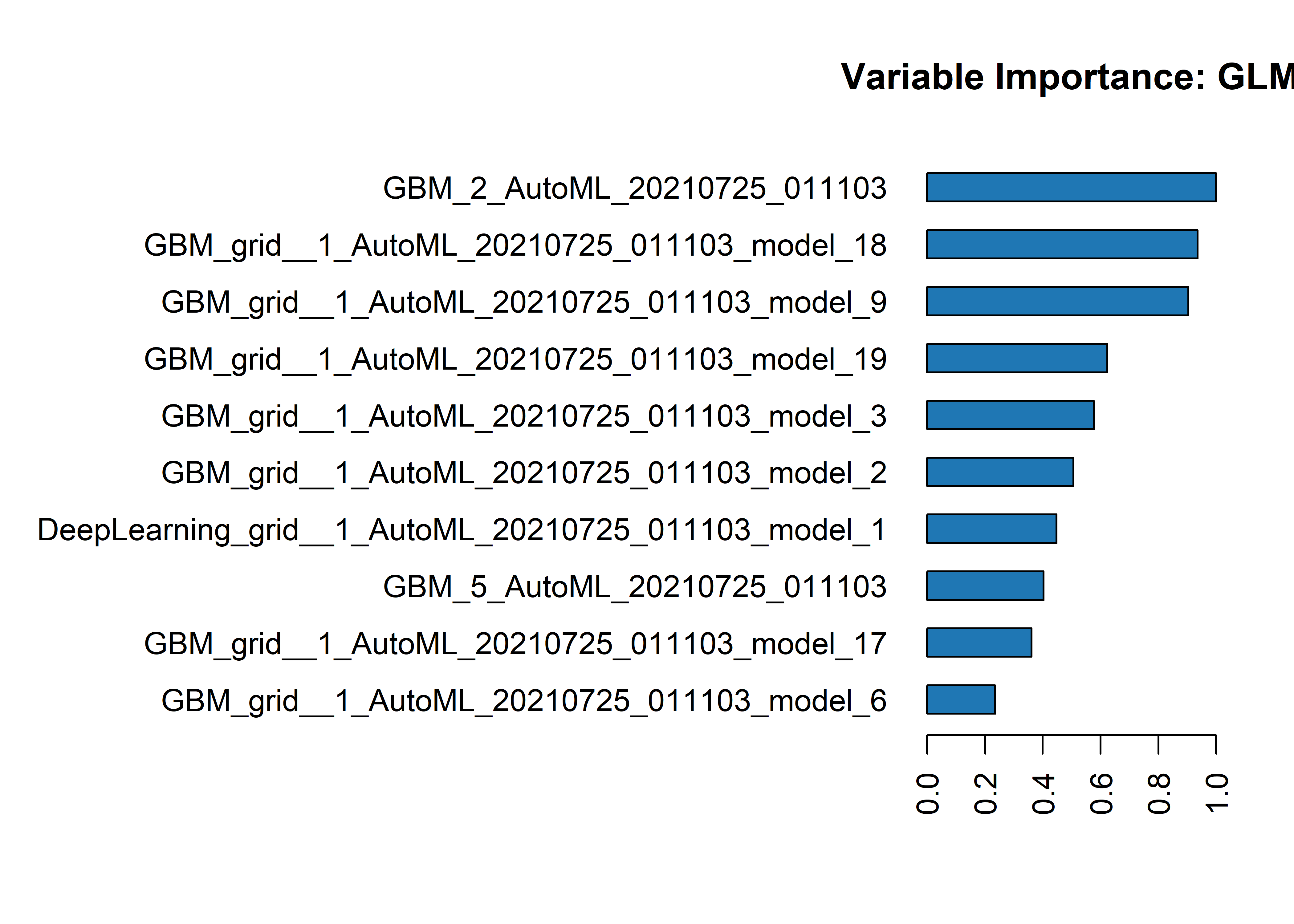
Importance of Features in the Original Dataset
Now let’s dig into the best individual model a bit to understand the parameters and feature importance of the original dataset. The top individual model is extracted and the variable importance can be displayed just like we did for the ensemble components.
top_individual_model <- h2o.getModel(metalearner_model@model$names[1])
metalearner_model@model$names[1]## [1] "GBM_2_AutoML_20210725_011103"h2o.varimp(top_individual_model) %>%
as_tibble() %>%
gt() %>%
cols_align("center")| variable | relative_importance | scaled_importance | percentage |
|---|---|---|---|
| total_trans_ct | 972.9781494 | 1.0000000000 | 0.2535628117 |
| total_trans_amt | 773.8638306 | 0.7953558166 | 0.2016726572 |
| total_ct_chng_q4_q1 | 567.8966675 | 0.5836684697 | 0.1479966183 |
| total_revolving_bal | 500.5438232 | 0.5144450814 | 0.1304441413 |
| total_relationship_count | 341.5622864 | 0.3510482600 | 0.0890127838 |
| total_amt_chng_q4_q1 | 175.8822327 | 0.1807668885 | 0.0458357605 |
| customer_age | 160.4104614 | 0.1648654305 | 0.0418037421 |
| avg_utilization_ratio | 132.2019196 | 0.1358734722 | 0.0344524597 |
| credit_limit | 72.8332520 | 0.0748559996 | 0.0189806977 |
| months_inactive_12_mon | 70.7272568 | 0.0726915161 | 0.0184318652 |
| gender_M | 37.5323524 | 0.0385747126 | 0.0097811126 |
| income_category_X.80K....120K | 6.2846680 | 0.0064592077 | 0.0016378149 |
| income_category_X.40K....60K | 6.1469312 | 0.0063176456 | 0.0016019200 |
| education_level_High.School | 3.8736489 | 0.0039812291 | 0.0010094916 |
| education_level_Graduate | 3.1741629 | 0.0032623167 | 0.0008272022 |
| education_level_Unknown | 2.5144935 | 0.0025843268 | 0.0006552892 |
| income_category_Less.than..40K | 1.8054181 | 0.0018555588 | 0.0004705007 |
| education_level_Uneducated | 1.7907501 | 0.0018404834 | 0.0004666781 |
| income_category_Unknown | 1.4599973 | 0.0015005448 | 0.0003804824 |
| income_category_X.60K....80K | 1.4241761 | 0.0014637288 | 0.0003711472 |
| education_level_Post.Graduate | 1.4217629 | 0.0014612486 | 0.0003705183 |
| education_level_Doctorate | 0.8990833 | 0.0009240529 | 0.0002343054 |
h2o.varimp_plot(top_individual_model)
If we were interested in the hyperparameters of this individual GB model, we could look at them like this:
top_individual_model@parameters## $model_id
## [1] "GBM_2_AutoML_20210725_011103"
##
## $training_frame
## [1] "automl_training_baked_dataset_tbl_sid_914f_1"
##
## $nfolds
## [1] 5
##
## $keep_cross_validation_models
## [1] FALSE
##
## $keep_cross_validation_predictions
## [1] TRUE
##
## $score_tree_interval
## [1] 5
##
## $fold_assignment
## [1] "Modulo"
##
## $ntrees
## [1] 101
##
## $max_depth
## [1] 7
##
## $stopping_metric
## [1] "logloss"
##
## $stopping_tolerance
## [1] 0.01187786
##
## $seed
## [1] 7252024
##
## $distribution
## [1] "bernoulli"
##
## $sample_rate
## [1] 0.8
##
## $col_sample_rate
## [1] 0.8
##
## $col_sample_rate_per_tree
## [1] 0.8
##
## $histogram_type
## [1] "UniformAdaptive"
##
## $categorical_encoding
## [1] "Enum"
##
## $x
## [1] "customer_age" "total_relationship_count"
## [3] "months_inactive_12_mon" "credit_limit"
## [5] "total_revolving_bal" "total_amt_chng_q4_q1"
## [7] "total_trans_amt" "total_trans_ct"
## [9] "total_ct_chng_q4_q1" "avg_utilization_ratio"
## [11] "gender_M" "education_level_Doctorate"
## [13] "education_level_Graduate" "education_level_High.School"
## [15] "education_level_Post.Graduate" "education_level_Uneducated"
## [17] "education_level_Unknown" "income_category_X.40K....60K"
## [19] "income_category_X.60K....80K" "income_category_X.80K....120K"
## [21] "income_category_Less.than..40K" "income_category_Unknown"
##
## $y
## [1] "churned"Final Thoughts
My takeaway is that h2o autoML is really quite powerful for purely predictive tasks and is even feasible for a timed competition like Sliced. It would not have won the modeling portion of Sliced, but still did well. There are also other aspects of winning the competition that I ignored (Data Vis, Golden Features).
I look forward to the upcoming playoffs and will be interested to try h2o on some new datasets to see if we just got lucky here, or if it’s really that good.
TLDR
AutoML wouldn’t have won this competition (at least with the minimal feature engineering I did), but it sure got way closer than I expected!
Thank you for your attention!
Session Info
sessionInfo()## R version 4.0.3 (2020-10-10)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 18363)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United States.1252
## [2] LC_CTYPE=English_United States.1252
## [3] LC_MONETARY=English_United States.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] gt_0.2.2 here_1.0.0 h2o_3.32.1.3 forcats_0.5.0
## [5] stringr_1.4.0 readr_1.4.0 tidyverse_1.3.0 yardstick_0.0.8
## [9] workflowsets_0.0.2 workflows_0.2.3 tune_0.1.5 tidyr_1.1.3
## [13] tibble_3.1.2 rsample_0.1.0 recipes_0.1.16 purrr_0.3.4
## [17] parsnip_0.1.6.9000 modeldata_0.1.0 infer_0.5.4 ggplot2_3.3.5
## [21] dplyr_1.0.7 dials_0.0.9 scales_1.1.1 broom_0.7.8
## [25] tidymodels_0.1.3
##
## loaded via a namespace (and not attached):
## [1] colorspace_2.0-0 ellipsis_0.3.2 class_7.3-17 rprojroot_2.0.2
## [5] fs_1.5.0 rstudioapi_0.13 listenv_0.8.0 furrr_0.2.1
## [9] bit64_4.0.5 prodlim_2019.11.13 fansi_0.5.0 lubridate_1.7.9.2
## [13] xml2_1.3.2 codetools_0.2-18 splines_4.0.3 knitr_1.30
## [17] jsonlite_1.7.1 pROC_1.16.2 dbplyr_2.0.0 compiler_4.0.3
## [21] httr_1.4.2 backports_1.2.0 assertthat_0.2.1 Matrix_1.2-18
## [25] cli_3.0.1 htmltools_0.5.0 tools_4.0.3 gtable_0.3.0
## [29] glue_1.4.2 Rcpp_1.0.5 cellranger_1.1.0 DiceDesign_1.8-1
## [33] vctrs_0.3.8 blogdown_0.15 iterators_1.0.13 timeDate_3043.102
## [37] gower_0.2.2 xfun_0.19 globals_0.14.0 rvest_0.3.6
## [41] lifecycle_1.0.0 future_1.20.1 MASS_7.3-53 ipred_0.9-9
## [45] hms_0.5.3 parallel_4.0.3 yaml_2.2.1 sass_0.3.1
## [49] rpart_4.1-15 stringi_1.5.3 foreach_1.5.1 checkmate_2.0.0
## [53] lhs_1.1.1 hardhat_0.1.6 lava_1.6.8.1 rlang_0.4.11
## [57] pkgconfig_2.0.3 bitops_1.0-6 evaluate_0.14 lattice_0.20-41
## [61] bit_4.0.4 tidyselect_1.1.1 parallelly_1.21.0 plyr_1.8.6
## [65] magrittr_2.0.1 bookdown_0.21 R6_2.5.0 generics_0.1.0
## [69] DBI_1.1.0 pillar_1.6.1 haven_2.3.1 withr_2.3.0
## [73] survival_3.2-7 RCurl_1.98-1.2 nnet_7.3-14 modelr_0.1.8
## [77] crayon_1.4.1 utf8_1.2.1 rmarkdown_2.5 grid_4.0.3
## [81] readxl_1.3.1 data.table_1.14.0 reprex_0.3.0 digest_0.6.27
## [85] GPfit_1.0-8 munsell_0.5.0Appendix - Full Details of the Ensemble Model
str(metalearner_model@model)## List of 35
## $ names : chr [1:37] "GBM_2_AutoML_20210725_011103" "GBM_grid__1_AutoML_20210725_011103_model_2" "GBM_4_AutoML_20210725_011103" "GBM_grid__1_AutoML_20210725_011103_model_11" ...
## $ original_names : NULL
## $ column_types : chr [1:37] "Numeric" "Numeric" "Numeric" "Numeric" ...
## $ domains :List of 37
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : NULL
## ..$ : chr [1:2] "no" "yes"
## $ cross_validation_models : NULL
## $ cross_validation_predictions : NULL
## $ cross_validation_holdout_predictions_frame_id : NULL
## $ cross_validation_fold_assignment_frame_id : NULL
## $ model_summary :Classes 'H2OTable' and 'data.frame': 1 obs. of 7 variables:
## ..$ family : chr "binomial"
## ..$ link : chr "logit"
## ..$ regularization : chr "Elastic Net (alpha = 0.5, lambda = 0.00311 )"
## ..$ number_of_predictors_total : int 36
## ..$ number_of_active_predictors: int 12
## ..$ number_of_iterations : int 7
## ..$ training_frame : chr "levelone_training_StackedEnsemble_AllModels_AutoML_20210725_011103"
## ..- attr(*, "header")= chr "GLM Model"
## ..- attr(*, "formats")= chr [1:7] "%s" "%s" "%s" "%d" ...
## ..- attr(*, "description")= chr "summary"
## $ scoring_history :Classes 'H2OTable' and 'data.frame': 2 obs. of 17 variables:
## ..$ timestamp : chr [1:2] "2021-07-25 01:26:33" "2021-07-25 01:26:33"
## ..$ duration : chr [1:2] " 0.000 sec" " 0.172 sec"
## ..$ iterations : int [1:2] 5 7
## ..$ negative_log_likelihood : num [1:2] 571 571
## ..$ objective : num [1:2] 0.0822 0.0837
## ..$ alpha : num [1:2] 0.5 1
## ..$ lambda : num [1:2] 0.00311 0.00311
## ..$ deviance_train : num [1:2] 0.161 0.161
## ..$ deviance_xval : num [1:2] 0.166 0.166
## ..$ deviance_se : num [1:2] 0.075 0.0751
## ..$ training_rmse : num [1:2] 0.152 0.152
## ..$ training_logloss : num [1:2] 0.0806 0.0805
## ..$ training_r2 : num [1:2] 0.828 0.828
## ..$ training_auc : num [1:2] 0.992 0.992
## ..$ training_pr_auc : num [1:2] 0.966 0.965
## ..$ training_lift : num [1:2] 6.26 6.26
## ..$ training_classification_error: num [1:2] 0.0295 0.0298
## ..- attr(*, "header")= chr "Scoring History"
## ..- attr(*, "formats")= chr [1:17] "%s" "%s" "%d" "%.5f" ...
## ..- attr(*, "description")= chr ""
## $ cv_scoring_history :List of 5
## ..$ :Classes 'H2OTable' and 'data.frame': 2 obs. of 23 variables:
## .. ..$ timestamp : chr [1:2] "2021-07-25 01:26:31" "2021-07-25 01:26:31"
## .. ..$ duration : chr [1:2] " 0.000 sec" " 0.205 sec"
## .. ..$ iterations : int [1:2] 5 7
## .. ..$ negative_log_likelihood : num [1:2] 473 473
## .. ..$ objective : num [1:2] 0.0684 0.0698
## .. ..$ alpha : num [1:2] 0.5 1
## .. ..$ lambda : num [1:2] 0.00311 0.00311
## .. ..$ deviance_train : num [1:2] 0.166 0.166
## .. ..$ deviance_test : num [1:2] 0.144 0.144
## .. ..$ training_rmse : num [1:2] 0.154 0.154
## .. ..$ training_logloss : num [1:2] 0.083 0.083
## .. ..$ training_r2 : num [1:2] 0.826 0.826
## .. ..$ training_auc : num [1:2] 0.992 0.992
## .. ..$ training_pr_auc : num [1:2] 0.964 0.964
## .. ..$ training_lift : num [1:2] 6.16 6.16
## .. ..$ training_classification_error : num [1:2] 0.0297 0.0295
## .. ..$ validation_rmse : num [1:2] 0.147 0.147
## .. ..$ validation_logloss : num [1:2] 0.072 0.0721
## .. ..$ validation_r2 : num [1:2] 0.83 0.829
## .. ..$ validation_auc : num [1:2] 0.994 0.994
## .. ..$ validation_pr_auc : num [1:2] 0.97 0.97
## .. ..$ validation_lift : num [1:2] 6.71 6.71
## .. ..$ validation_classification_error: num [1:2] 0.0288 0.0295
## .. ..- attr(*, "header")= chr "Scoring History"
## .. ..- attr(*, "formats")= chr [1:23] "%s" "%s" "%d" "%.5f" ...
## .. ..- attr(*, "description")= chr ""
## ..$ :Classes 'H2OTable' and 'data.frame': 2 obs. of 23 variables:
## .. ..$ timestamp : chr [1:2] "2021-07-25 01:26:31" "2021-07-25 01:26:32"
## .. ..$ duration : chr [1:2] " 0.000 sec" " 0.192 sec"
## .. ..$ iterations : int [1:2] 5 7
## .. ..$ negative_log_likelihood : num [1:2] 448 448
## .. ..$ objective : num [1:2] 0.065 0.0664
## .. ..$ alpha : num [1:2] 0.5 1
## .. ..$ lambda : num [1:2] 0.00311 0.00311
## .. ..$ deviance_train : num [1:2] 0.157 0.157
## .. ..$ deviance_test : num [1:2] 0.179 0.179
## .. ..$ training_rmse : num [1:2] 0.151 0.151
## .. ..$ training_logloss : num [1:2] 0.0785 0.0785
## .. ..$ training_r2 : num [1:2] 0.831 0.831
## .. ..$ training_auc : num [1:2] 0.993 0.993
## .. ..$ training_pr_auc : num [1:2] 0.968 0.968
## .. ..$ training_lift : num [1:2] 6.21 6.21
## .. ..$ training_classification_error : num [1:2] 0.0293 0.0293
## .. ..$ validation_rmse : num [1:2] 0.157 0.157
## .. ..$ validation_logloss : num [1:2] 0.0894 0.0894
## .. ..$ validation_r2 : num [1:2] 0.811 0.812
## .. ..$ validation_auc : num [1:2] 0.99 0.99
## .. ..$ validation_pr_auc : num [1:2] 0.954 0.954
## .. ..$ validation_lift : num [1:2] 6.5 6.5
## .. ..$ validation_classification_error: num [1:2] 0.0305 0.0305
## .. ..- attr(*, "header")= chr "Scoring History"
## .. ..- attr(*, "formats")= chr [1:23] "%s" "%s" "%d" "%.5f" ...
## .. ..- attr(*, "description")= chr ""
## ..$ :Classes 'H2OTable' and 'data.frame': 2 obs. of 23 variables:
## .. ..$ timestamp : chr [1:2] "2021-07-25 01:26:32" "2021-07-25 01:26:32"
## .. ..$ duration : chr [1:2] " 0.000 sec" " 0.208 sec"
## .. ..$ iterations : int [1:2] 5 7
## .. ..$ negative_log_likelihood : num [1:2] 440 440
## .. ..$ objective : num [1:2] 0.0638 0.0653
## .. ..$ alpha : num [1:2] 0.5 1
## .. ..$ lambda : num [1:2] 0.00311 0.00311
## .. ..$ deviance_train : num [1:2] 0.155 0.155
## .. ..$ deviance_test : num [1:2] 0.19 0.191
## .. ..$ training_rmse : num [1:2] 0.149 0.149
## .. ..$ training_logloss : num [1:2] 0.0774 0.0773
## .. ..$ training_r2 : num [1:2] 0.832 0.832
## .. ..$ training_auc : num [1:2] 0.993 0.993
## .. ..$ training_pr_auc : num [1:2] 0.967 0.967
## .. ..$ training_lift : num [1:2] 6.37 6.37
## .. ..$ training_classification_error : num [1:2] 0.0294 0.0294
## .. ..$ validation_rmse : num [1:2] 0.166 0.166
## .. ..$ validation_logloss : num [1:2] 0.0948 0.0953
## .. ..$ validation_r2 : num [1:2] 0.806 0.805
## .. ..$ validation_auc : num [1:2] 0.99 0.99
## .. ..$ validation_pr_auc : num [1:2] 0.959 0.959
## .. ..$ validation_lift : num [1:2] 5.86 5.86
## .. ..$ validation_classification_error: num [1:2] 0.0314 0.0321
## .. ..- attr(*, "header")= chr "Scoring History"
## .. ..- attr(*, "formats")= chr [1:23] "%s" "%s" "%d" "%.5f" ...
## .. ..- attr(*, "description")= chr ""
## ..$ :Classes 'H2OTable' and 'data.frame': 2 obs. of 23 variables:
## .. ..$ timestamp : chr [1:2] "2021-07-25 01:26:32" "2021-07-25 01:26:32"
## .. ..$ duration : chr [1:2] " 0.000 sec" " 0.173 sec"
## .. ..$ iterations : int [1:2] 5 7
## .. ..$ negative_log_likelihood : num [1:2] 470 470
## .. ..$ objective : num [1:2] 0.0679 0.0694
## .. ..$ alpha : num [1:2] 0.5 1
## .. ..$ lambda : num [1:2] 0.00311 0.00311
## .. ..$ deviance_train : num [1:2] 0.168 0.169
## .. ..$ deviance_test : num [1:2] 0.137 0.136
## .. ..$ training_rmse : num [1:2] 0.156 0.156
## .. ..$ training_logloss : num [1:2] 0.0842 0.0842
## .. ..$ training_r2 : num [1:2] 0.82 0.82
## .. ..$ training_auc : num [1:2] 0.992 0.992
## .. ..$ training_pr_auc : num [1:2] 0.963 0.963
## .. ..$ training_lift : num [1:2] 6.17 6.17
## .. ..$ training_classification_error : num [1:2] 0.0317 0.0314
## .. ..$ validation_rmse : num [1:2] 0.136 0.136
## .. ..$ validation_logloss : num [1:2] 0.0683 0.0682
## .. ..$ validation_r2 : num [1:2] 0.856 0.856
## .. ..$ validation_auc : num [1:2] 0.994 0.994
## .. ..$ validation_pr_auc : num [1:2] 0.972 0.972
## .. ..$ validation_lift : num [1:2] 6.61 6.61
## .. ..$ validation_classification_error: num [1:2] 0.0206 0.0206
## .. ..- attr(*, "header")= chr "Scoring History"
## .. ..- attr(*, "formats")= chr [1:23] "%s" "%s" "%d" "%.5f" ...
## .. ..- attr(*, "description")= chr ""
## ..$ :Classes 'H2OTable' and 'data.frame': 2 obs. of 23 variables:
## .. ..$ timestamp : chr [1:2] "2021-07-25 01:26:32" "2021-07-25 01:26:33"
## .. ..$ duration : chr [1:2] " 0.000 sec" " 0.179 sec"
## .. ..$ iterations : int [1:2] 5 7
## .. ..$ negative_log_likelihood : num [1:2] 445 445
## .. ..$ objective : num [1:2] 0.0645 0.0659
## .. ..$ alpha : num [1:2] 0.5 1
## .. ..$ lambda : num [1:2] 0.00311 0.00311
## .. ..$ deviance_train : num [1:2] 0.157 0.157
## .. ..$ deviance_test : num [1:2] 0.181 0.181
## .. ..$ training_rmse : num [1:2] 0.149 0.149
## .. ..$ training_logloss : num [1:2] 0.0784 0.0784
## .. ..$ training_r2 : num [1:2] 0.832 0.832
## .. ..$ training_auc : num [1:2] 0.992 0.992
## .. ..$ training_pr_auc : num [1:2] 0.966 0.966
## .. ..$ training_lift : num [1:2] 6.41 6.41
## .. ..$ training_classification_error : num [1:2] 0.0275 0.0275
## .. ..$ validation_rmse : num [1:2] 0.165 0.165
## .. ..$ validation_logloss : num [1:2] 0.0905 0.0903
## .. ..$ validation_r2 : num [1:2] 0.811 0.811
## .. ..$ validation_auc : num [1:2] 0.991 0.992
## .. ..$ validation_pr_auc : num [1:2] 0.964 0.964
## .. ..$ validation_lift : num [1:2] 5.74 5.74
## .. ..$ validation_classification_error: num [1:2] 0.0361 0.0369
## .. ..- attr(*, "header")= chr "Scoring History"
## .. ..- attr(*, "formats")= chr [1:23] "%s" "%s" "%d" "%.5f" ...
## .. ..- attr(*, "description")= chr ""
## $ reproducibility_information_table :List of 3
## ..$ :Classes 'H2OTable' and 'data.frame': 1 obs. of 26 variables:
## .. ..$ node : int 0
## .. ..$ h2o : chr "127.0.0.1:54321"
## .. ..$ healthy : chr "true"
## .. ..$ last_ping : chr "1627201591256"
## .. ..$ num_cpus : int 4
## .. ..$ sys_load : num 0.564
## .. ..$ mem_value_size : num 70732923
## .. ..$ free_mem : num 7.76e+08
## .. ..$ pojo_mem : num 1.91e+08
## .. ..$ swap_mem : num 0
## .. ..$ free_disc : num 3.6e+10
## .. ..$ max_disc : num 2.55e+11
## .. ..$ pid : int 7916
## .. ..$ num_keys : int 9133
## .. ..$ tcps_active : chr ""
## .. ..$ open_fds : int -1
## .. ..$ rpcs_active : chr ""
## .. ..$ nthreads : int 4
## .. ..$ is_leader : chr "true"
## .. ..$ total_mem : num 6.27e+08
## .. ..$ max_mem : num 1.04e+09
## .. ..$ java_version : chr "Java 1.8.0_77 (from Oracle Corporation)"
## .. ..$ jvm_launch_parameters: chr "[-Xmx1g, -ea]"
## .. ..$ os_version : chr "Windows 10 10.0 (x86)"
## .. ..$ machine_physical_mem : num 1.7e+10
## .. ..$ machine_locale : chr "en_US"
## .. ..- attr(*, "header")= chr "Node Information"
## .. ..- attr(*, "formats")= chr [1:26] "%d" "%s" "%s" "%s" ...
## .. ..- attr(*, "description")= chr ""
## ..$ :Classes 'H2OTable' and 'data.frame': 1 obs. of 13 variables:
## .. ..$ h2o_cluster_uptime : num 1394453
## .. ..$ h2o_cluster_timezone : chr "America/Los_Angeles"
## .. ..$ h2o_data_parsing_timezone: chr "UTC"
## .. ..$ h2o_cluster_version : chr "3.32.1.3"
## .. ..$ h2o_cluster_version_age : chr "2 months and 5 days"
## .. ..$ h2o_cluster_name : chr "H2O_started_from_R_kingr17_afp345"
## .. ..$ h2o_cluster_total_nodes : int 1
## .. ..$ h2o_cluster_free_memory : num 7.76e+08
## .. ..$ h2o_cluster_total_cores : int 4
## .. ..$ h2o_cluster_allowed_cores: int 4
## .. ..$ h2o_cluster_status : chr "locked, healthly"
## .. ..$ h2o_internal_security : chr "false"
## .. ..$ h2o_api_extensions : chr "Amazon S3, Algos, AutoML, Core V3, TargetEncoder, Core V4"
## .. ..- attr(*, "header")= chr "Cluster Configuration"
## .. ..- attr(*, "formats")= chr [1:13] "%d" "%s" "%s" "%s" ...
## .. ..- attr(*, "description")= chr ""
## ..$ :Classes 'H2OTable' and 'data.frame': 2 obs. of 3 variables:
## .. ..$ input_frame: chr [1:2] "training_frame" "validation_frame"
## .. ..$ checksum : num [1:2] 3.75e+18 -1.00
## .. ..$ espc : chr [1:2] "[0, 1772, 3544, 5316, 7088]" "-1"
## .. ..- attr(*, "header")= chr "Input Frames Information"
## .. ..- attr(*, "formats")= chr [1:3] "%s" "%d" "%d"
## .. ..- attr(*, "description")= chr ""
## $ training_metrics :Formal class 'H2OBinomialMetrics' [package "h2o"] with 5 slots
## .. ..@ algorithm: chr "glm"
## .. ..@ on_train : logi TRUE
## .. ..@ on_valid : logi FALSE
## .. ..@ on_xval : logi FALSE
## .. ..@ metrics :List of 30
## .. .. ..$ __meta :List of 3
## .. .. .. ..$ schema_version: int 3
## .. .. .. ..$ schema_name : chr "ModelMetricsBinomialGLMV3"
## .. .. .. ..$ schema_type : chr "ModelMetricsBinomialGLM"
## .. .. ..$ model :List of 4
## .. .. .. ..$ __meta:List of 3
## .. .. .. .. ..$ schema_version: int 3
## .. .. .. .. ..$ schema_name : chr "ModelKeyV3"
## .. .. .. .. ..$ schema_type : chr "Key<Model>"
## .. .. .. ..$ name : chr "metalearner_AUTO_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. .. ..$ type : chr "Key<Model>"
## .. .. .. ..$ URL : chr "/3/Models/metalearner_AUTO_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. ..$ model_checksum : chr "1672530778380092240"
## .. .. ..$ frame :List of 4
## .. .. .. ..$ __meta:List of 3
## .. .. .. .. ..$ schema_version: int 3
## .. .. .. .. ..$ schema_name : chr "FrameKeyV3"
## .. .. .. .. ..$ schema_type : chr "Key<Frame>"
## .. .. .. ..$ name : chr "levelone_training_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. .. ..$ type : chr "Key<Frame>"
## .. .. .. ..$ URL : chr "/3/Frames/levelone_training_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. ..$ frame_checksum : chr "3754607664224636173"
## .. .. ..$ description : NULL
## .. .. ..$ model_category : chr "Binomial"
## .. .. ..$ scoring_time : num 1.63e+12
## .. .. ..$ predictions : NULL
## .. .. ..$ MSE : num 0.0231
## .. .. ..$ RMSE : num 0.152
## .. .. ..$ nobs : int 7088
## .. .. ..$ custom_metric_name : NULL
## .. .. ..$ custom_metric_value : num 0
## .. .. ..$ r2 : num 0.828
## .. .. ..$ logloss : num 0.0805
## .. .. ..$ AUC : num 0.992
## .. .. ..$ pr_auc : num 0.965
## .. .. ..$ Gini : num 0.985
## .. .. ..$ mean_per_class_error : num 0.0563
## .. .. ..$ domain : chr [1:2] "no" "yes"
## .. .. ..$ cm :List of 2
## .. .. .. ..$ __meta:List of 3
## .. .. .. .. ..$ schema_version: int 3
## .. .. .. .. ..$ schema_name : chr "ConfusionMatrixV3"
## .. .. .. .. ..$ schema_type : chr "ConfusionMatrix"
## .. .. .. ..$ table :Classes 'H2OTable' and 'data.frame': 3 obs. of 4 variables:
## .. .. .. .. ..$ no : num [1:3] 5853 108 5961
## .. .. .. .. ..$ yes : num [1:3] 103 1024 1127
## .. .. .. .. ..$ Error: num [1:3] 0.0173 0.0954 0.0298
## .. .. .. .. ..$ Rate : chr [1:3] "103 / 5,956" "108 / 1,132" "211 / 7,088"
## .. .. .. .. ..- attr(*, "header")= chr "Confusion Matrix"
## .. .. .. .. ..- attr(*, "formats")= chr [1:4] "%d" "%d" "%.4f" "= %11s"
## .. .. .. .. ..- attr(*, "description")= chr "Row labels: Actual class; Column labels: Predicted class"
## .. .. ..$ thresholds_and_metric_scores :Classes 'H2OTable' and 'data.frame': 400 obs. of 20 variables:
## .. .. .. ..$ threshold : num [1:400] 0.999 0.999 0.998 0.998 0.997 ...
## .. .. .. ..$ f1 : num [1:400] 0.0666 0.1305 0.1757 0.2299 0.2665 ...
## .. .. .. ..$ f2 : num [1:400] 0.0427 0.0857 0.1175 0.1572 0.185 ...
## .. .. .. ..$ f0point5 : num [1:400] 0.151 0.273 0.348 0.427 0.476 ...
## .. .. .. ..$ accuracy : num [1:400] 0.846 0.851 0.856 0.861 0.865 ...
## .. .. .. ..$ precision : num [1:400] 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ recall : num [1:400] 0.0345 0.0698 0.0963 0.1299 0.1537 ...
## .. .. .. ..$ specificity : num [1:400] 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ absolute_mcc : num [1:400] 0.171 0.244 0.287 0.334 0.364 ...
## .. .. .. ..$ min_per_class_accuracy : num [1:400] 0.0345 0.0698 0.0963 0.1299 0.1537 ...
## .. .. .. ..$ mean_per_class_accuracy: num [1:400] 0.517 0.535 0.548 0.565 0.577 ...
## .. .. .. ..$ tns : num [1:400] 5956 5956 5956 5956 5956 ...
## .. .. .. ..$ fns : num [1:400] 1093 1053 1023 985 958 ...
## .. .. .. ..$ fps : num [1:400] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. ..$ tps : num [1:400] 39 79 109 147 174 198 226 254 276 302 ...
## .. .. .. ..$ tnr : num [1:400] 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ fnr : num [1:400] 0.966 0.93 0.904 0.87 0.846 ...
## .. .. .. ..$ fpr : num [1:400] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. ..$ tpr : num [1:400] 0.0345 0.0698 0.0963 0.1299 0.1537 ...
## .. .. .. ..$ idx : int [1:400] 0 1 2 3 4 5 6 7 8 9 ...
## .. .. .. ..- attr(*, "header")= chr "Metrics for Thresholds"
## .. .. .. ..- attr(*, "formats")= chr [1:20] "%f" "%f" "%f" "%f" ...
## .. .. .. ..- attr(*, "description")= chr "Binomial metrics as a function of classification thresholds"
## .. .. ..$ max_criteria_and_metric_scores:Classes 'H2OTable' and 'data.frame': 18 obs. of 4 variables:
## .. .. .. ..$ metric : chr [1:18] "max f1" "max f2" "max f0point5" "max accuracy" ...
## .. .. .. ..$ threshold: num [1:18] 0.441 0.244 0.731 0.492 0.999 ...
## .. .. .. ..$ value : num [1:18] 0.907 0.923 0.926 0.97 1 ...
## .. .. .. ..$ idx : num [1:18] 191 243 119 180 0 378 0 191 272 254 ...
## .. .. .. ..- attr(*, "header")= chr "Maximum Metrics"
## .. .. .. ..- attr(*, "formats")= chr [1:4] "%s" "%f" "%f" "%d"
## .. .. .. ..- attr(*, "description")= chr "Maximum metrics at their respective thresholds"
## .. .. ..$ gains_lift_table :Classes 'H2OTable' and 'data.frame': 16 obs. of 14 variables:
## .. .. .. ..$ group : int [1:16] 1 2 3 4 5 6 7 8 9 10 ...
## .. .. .. ..$ cumulative_data_fraction: num [1:16] 0.01 0.02 0.0301 0.0401 0.0501 ...
## .. .. .. ..$ lower_threshold : num [1:16] 0.999 0.998 0.996 0.995 0.993 ...
## .. .. .. ..$ lift : num [1:16] 6.26 6.26 6.26 6.26 6.26 ...
## .. .. .. ..$ cumulative_lift : num [1:16] 6.26 6.26 6.26 6.26 6.26 ...
## .. .. .. ..$ response_rate : num [1:16] 1 1 1 1 1 ...
## .. .. .. ..$ score : num [1:16] 0.999 0.998 0.997 0.996 0.994 ...
## .. .. .. ..$ cumulative_response_rate: num [1:16] 1 1 1 1 1 ...
## .. .. .. ..$ cumulative_score : num [1:16] 0.999 0.999 0.998 0.997 0.997 ...
## .. .. .. ..$ capture_rate : num [1:16] 0.0627 0.0627 0.0627 0.0627 0.0627 ...
## .. .. .. ..$ cumulative_capture_rate : num [1:16] 0.0627 0.1254 0.1882 0.2509 0.3136 ...
## .. .. .. ..$ gain : num [1:16] 526 526 526 526 526 ...
## .. .. .. ..$ cumulative_gain : num [1:16] 526 526 526 526 526 ...
## .. .. .. ..$ kolmogorov_smirnov : num [1:16] 0.0627 0.1254 0.1882 0.2509 0.3136 ...
## .. .. .. ..- attr(*, "header")= chr "Gains/Lift Table"
## .. .. .. ..- attr(*, "formats")= chr [1:14] "%d" "%.8f" "%5f" "%5f" ...
## .. .. .. ..- attr(*, "description")= chr "Avg response rate: 15.97 %, avg score: 15.97 %"
## .. .. ..$ residual_deviance : num 1141
## .. .. ..$ null_deviance : num 6226
## .. .. ..$ AIC : num 1167
## .. .. ..$ null_degrees_of_freedom : int 7087
## .. .. ..$ residual_degrees_of_freedom : int 7075
## $ validation_metrics :Formal class 'H2OBinomialMetrics' [package "h2o"] with 5 slots
## .. ..@ algorithm: chr "glm"
## .. ..@ on_train : logi FALSE
## .. ..@ on_valid : logi TRUE
## .. ..@ on_xval : logi FALSE
## .. ..@ metrics : NULL
## $ cross_validation_metrics :Formal class 'H2OBinomialMetrics' [package "h2o"] with 5 slots
## .. ..@ algorithm: chr "glm"
## .. ..@ on_train : logi FALSE
## .. ..@ on_valid : logi FALSE
## .. ..@ on_xval : logi TRUE
## .. ..@ metrics :List of 30
## .. .. ..$ __meta :List of 3
## .. .. .. ..$ schema_version: int 3
## .. .. .. ..$ schema_name : chr "ModelMetricsBinomialGLMV3"
## .. .. .. ..$ schema_type : chr "ModelMetricsBinomialGLM"
## .. .. ..$ model :List of 4
## .. .. .. ..$ __meta:List of 3
## .. .. .. .. ..$ schema_version: int 3
## .. .. .. .. ..$ schema_name : chr "ModelKeyV3"
## .. .. .. .. ..$ schema_type : chr "Key<Model>"
## .. .. .. ..$ name : chr "metalearner_AUTO_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. .. ..$ type : chr "Key<Model>"
## .. .. .. ..$ URL : chr "/3/Models/metalearner_AUTO_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. ..$ model_checksum : chr "1672530778380092240"
## .. .. ..$ frame :List of 4
## .. .. .. ..$ __meta:List of 3
## .. .. .. .. ..$ schema_version: int 3
## .. .. .. .. ..$ schema_name : chr "FrameKeyV3"
## .. .. .. .. ..$ schema_type : chr "Key<Frame>"
## .. .. .. ..$ name : chr "levelone_training_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. .. ..$ type : chr "Key<Frame>"
## .. .. .. ..$ URL : chr "/3/Frames/levelone_training_StackedEnsemble_AllModels_AutoML_20210725_011103"
## .. .. ..$ frame_checksum : chr "3754607664224636173"
## .. .. ..$ description : chr "5-fold cross-validation on training data (Metrics computed for combined holdout predictions)"
## .. .. ..$ model_category : chr "Binomial"
## .. .. ..$ scoring_time : num 1.63e+12
## .. .. ..$ predictions : NULL
## .. .. ..$ MSE : num 0.0238
## .. .. ..$ RMSE : num 0.154
## .. .. ..$ nobs : int 7088
## .. .. ..$ custom_metric_name : NULL
## .. .. ..$ custom_metric_value : num 0
## .. .. ..$ r2 : num 0.823
## .. .. ..$ logloss : num 0.0829
## .. .. ..$ AUC : num 0.992
## .. .. ..$ pr_auc : num 0.964
## .. .. ..$ Gini : num 0.984
## .. .. ..$ mean_per_class_error : num 0.0601
## .. .. ..$ domain : chr [1:2] "no" "yes"
## .. .. ..$ cm :List of 2
## .. .. .. ..$ __meta:List of 3
## .. .. .. .. ..$ schema_version: int 3
## .. .. .. .. ..$ schema_name : chr "ConfusionMatrixV3"
## .. .. .. .. ..$ schema_type : chr "ConfusionMatrix"
## .. .. .. ..$ table :Classes 'H2OTable' and 'data.frame': 3 obs. of 4 variables:
## .. .. .. .. ..$ no : num [1:3] 5856 117 5973
## .. .. .. .. ..$ yes : num [1:3] 100 1015 1115
## .. .. .. .. ..$ Error: num [1:3] 0.0168 0.1034 0.0306
## .. .. .. .. ..$ Rate : chr [1:3] "100 / 5,956" "117 / 1,132" "217 / 7,088"
## .. .. .. .. ..- attr(*, "header")= chr "Confusion Matrix"
## .. .. .. .. ..- attr(*, "formats")= chr [1:4] "%d" "%d" "%.4f" "= %11s"
## .. .. .. .. ..- attr(*, "description")= chr "Row labels: Actual class; Column labels: Predicted class"
## .. .. ..$ thresholds_and_metric_scores :Classes 'H2OTable' and 'data.frame': 400 obs. of 20 variables:
## .. .. .. ..$ threshold : num [1:400] 0.999 0.999 0.998 0.998 0.997 ...
## .. .. .. ..$ f1 : num [1:400] 0.0781 0.1258 0.1668 0.2046 0.2463 ...
## .. .. .. ..$ f2 : num [1:400] 0.0503 0.0825 0.1112 0.1385 0.1696 ...
## .. .. .. ..$ f0point5 : num [1:400] 0.175 0.265 0.334 0.391 0.45 ...
## .. .. .. ..$ accuracy : num [1:400] 0.847 0.851 0.855 0.858 0.863 ...
## .. .. .. ..$ precision : num [1:400] 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ recall : num [1:400] 0.0406 0.0671 0.091 0.114 0.1405 ...
## .. .. .. ..$ specificity : num [1:400] 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ absolute_mcc : num [1:400] 0.185 0.239 0.279 0.312 0.347 ...
## .. .. .. ..$ min_per_class_accuracy : num [1:400] 0.0406 0.0671 0.091 0.114 0.1405 ...
## .. .. .. ..$ mean_per_class_accuracy: num [1:400] 0.52 0.534 0.545 0.557 0.57 ...
## .. .. .. ..$ tns : num [1:400] 5956 5956 5956 5956 5956 ...
## .. .. .. ..$ fns : num [1:400] 1086 1056 1029 1003 973 ...
## .. .. .. ..$ fps : num [1:400] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. ..$ tps : num [1:400] 46 76 103 129 159 190 224 256 280 300 ...
## .. .. .. ..$ tnr : num [1:400] 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ fnr : num [1:400] 0.959 0.933 0.909 0.886 0.86 ...
## .. .. .. ..$ fpr : num [1:400] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. ..$ tpr : num [1:400] 0.0406 0.0671 0.091 0.114 0.1405 ...
## .. .. .. ..$ idx : int [1:400] 0 1 2 3 4 5 6 7 8 9 ...
## .. .. .. ..- attr(*, "header")= chr "Metrics for Thresholds"
## .. .. .. ..- attr(*, "formats")= chr [1:20] "%f" "%f" "%f" "%f" ...
## .. .. .. ..- attr(*, "description")= chr "Binomial metrics as a function of classification thresholds"
## .. .. ..$ max_criteria_and_metric_scores:Classes 'H2OTable' and 'data.frame': 18 obs. of 4 variables:
## .. .. .. ..$ metric : chr [1:18] "max f1" "max f2" "max f0point5" "max accuracy" ...
## .. .. .. ..$ threshold: num [1:18] 0.468 0.201 0.773 0.527 0.999 ...
## .. .. .. ..$ value : num [1:18] 0.903 0.921 0.924 0.97 1 ...
## .. .. .. ..$ idx : num [1:18] 183 256 108 168 0 381 0 183 272 258 ...
## .. .. .. ..- attr(*, "header")= chr "Maximum Metrics"
## .. .. .. ..- attr(*, "formats")= chr [1:4] "%s" "%f" "%f" "%d"
## .. .. .. ..- attr(*, "description")= chr "Maximum metrics at their respective thresholds"
## .. .. ..$ gains_lift_table :Classes 'H2OTable' and 'data.frame': 16 obs. of 14 variables:
## .. .. .. ..$ group : int [1:16] 1 2 3 4 5 6 7 8 9 10 ...
## .. .. .. ..$ cumulative_data_fraction: num [1:16] 0.01 0.02 0.0301 0.0401 0.0501 ...
## .. .. .. ..$ lower_threshold : num [1:16] 0.999 0.997 0.996 0.995 0.993 ...
## .. .. .. ..$ lift : num [1:16] 6.26 6.26 6.26 6.26 6.26 ...
## .. .. .. ..$ cumulative_lift : num [1:16] 6.26 6.26 6.26 6.26 6.26 ...
## .. .. .. ..$ response_rate : num [1:16] 1 1 1 1 1 ...
## .. .. .. ..$ score : num [1:16] 0.999 0.998 0.997 0.995 0.994 ...
## .. .. .. ..$ cumulative_response_rate: num [1:16] 1 1 1 1 1 ...
## .. .. .. ..$ cumulative_score : num [1:16] 0.999 0.999 0.998 0.997 0.997 ...
## .. .. .. ..$ capture_rate : num [1:16] 0.0627 0.0627 0.0627 0.0627 0.0627 ...
## .. .. .. ..$ cumulative_capture_rate : num [1:16] 0.0627 0.1254 0.1882 0.2509 0.3136 ...
## .. .. .. ..$ gain : num [1:16] 526 526 526 526 526 ...
## .. .. .. ..$ cumulative_gain : num [1:16] 526 526 526 526 526 ...
## .. .. .. ..$ kolmogorov_smirnov : num [1:16] 0.0627 0.1254 0.1882 0.2509 0.3136 ...
## .. .. .. ..- attr(*, "header")= chr "Gains/Lift Table"
## .. .. .. ..- attr(*, "formats")= chr [1:14] "%d" "%.8f" "%5f" "%5f" ...
## .. .. .. ..- attr(*, "description")= chr "Avg response rate: 15.97 %, avg score: 15.98 %"
## .. .. ..$ residual_deviance : num 1175
## .. .. ..$ null_deviance : num 6229
## .. .. ..$ AIC : num 1201
## .. .. ..$ null_degrees_of_freedom : int 7087
## .. .. ..$ residual_degrees_of_freedom : int 7075
## $ cross_validation_metrics_summary :Classes 'H2OTable' and 'data.frame': 22 obs. of 7 variables:
## ..$ mean : chr [1:22] "0.9701006" "0.9918038" "0.029899428" "42.2" ...
## ..$ sd : chr [1:22] "0.0059326575" "0.0018960662" "0.0059326575" "7.5960517" ...
## ..$ cv_1_valid: chr [1:22] "0.97050357" "0.993848" "0.029496403" "41.0" ...
## ..$ cv_2_valid: chr [1:22] "0.96954316" "0.9896141" "0.030456852" "42.0" ...
## ..$ cv_3_valid: chr [1:22] "0.9678801" "0.99040574" "0.032119915" "45.0" ...
## ..$ cv_4_valid: chr [1:22] "0.9794293" "0.9936388" "0.02057067" "31.0" ...
## ..$ cv_5_valid: chr [1:22] "0.9631467" "0.99151224" "0.036853295" "52.0" ...
## ..- attr(*, "header")= chr "Cross-Validation Metrics Summary"
## ..- attr(*, "formats")= chr [1:7] "%s" "%s" "%s" "%s" ...
## ..- attr(*, "description")= chr ""
## $ status : NULL
## $ start_time : num 1.63e+12
## $ end_time : num 1.63e+12
## $ run_time : int 266
## $ default_threshold : num 0.441
## $ help :List of 36
## ..$ domains : chr "Domains for categorical columns"
## ..$ __meta : chr "Metadata on this schema instance, to make it self-describing."
## ..$ standardized_coefficient_magnitudes : chr "Standardized Coefficient Magnitudes"
## ..$ help : chr "Help information for output fields"
## ..$ training_metrics : chr "Training data model metrics"
## ..$ cross_validation_models : chr "Cross-validation models (model ids)"
## ..$ cross_validation_predictions : chr "Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead)"
## ..$ lambda_best : chr "Lambda minimizing the objective value, only applicable with lambda search or when arrays of alpha and lambdas are provided"
## ..$ cross_validation_metrics_summary : chr "Cross-validation model metrics summary"
## ..$ status : chr "Job status"
## ..$ reproducibility_information_table : chr "Model reproducibility information"
## ..$ variable_importances : chr "Variable Importances"
## ..$ model_summary : chr "Model summary"
## ..$ end_time : chr "End time in milliseconds"
## ..$ names : chr "Column names"
## ..$ cross_validation_fold_assignment_frame_id : chr "Cross-validation fold assignment (each row is assigned to one holdout fold)"
## ..$ lambda_1se : chr "Lambda best + 1 standard error. Only applicable with lambda search and cross-validation"
## ..$ run_time : chr "Runtime in milliseconds"
## ..$ lambda_max : chr "Starting lambda value used when lambda search is enabled."
## ..$ default_threshold : chr "Default threshold used for predictions"
## ..$ scoring_history : chr "Scoring history"
## ..$ dispersion : chr "Dispersion parameter, only applicable to Tweedie family (input/output) and fractional Binomial (output only)"
## ..$ cross_validation_holdout_predictions_frame_id : chr "Cross-validation holdout predictions (full out-of-sample predictions on training data)"
## ..$ alpha_best : chr "Alpha minimizing the objective value, only applicable when arrays of alphas are given "
## ..$ lambda_min : chr "Minimum lambda value calculated that may be used for lambda search. Early-stop may happen and the minimum lamb"| __truncated__
## ..$ coefficients_table_multinomials_with_class_names: chr "Table of Coefficients with coefficients denoted with class names for GLM multinonimals only."
## ..$ model_category : chr "Category of the model (e.g., Binomial)"
## ..$ random_coefficients_table : chr "Table of Random Coefficients for HGLM"
## ..$ best_submodel_index : chr "submodel index minimizing the objective value, only applicable for arrays of alphas/lambda "
## ..$ column_types : chr "Column types"
## ..$ original_names : chr "Original column names"
## ..$ start_time : chr "Start time in milliseconds"
## ..$ cv_scoring_history : chr "Cross-Validation scoring history"
## ..$ cross_validation_metrics : chr "Cross-validation model metrics"
## ..$ coefficients_table : chr "Table of Coefficients"
## ..$ validation_metrics : chr "Validation data model metrics"
## $ coefficients_table :Classes 'H2OTable' and 'data.frame': 37 obs. of 3 variables:
## ..$ names : chr [1:37] "Intercept" "GBM_2_AutoML_20210725_011103" "GBM_grid__1_AutoML_20210725_011103_model_2" "GBM_4_AutoML_20210725_011103" ...
## ..$ coefficients : num [1:37] 0.3863 0.183 0.0834 0 0.0228 ...
## ..$ standardized_coefficients: num [1:37] -4.7185 0.7014 0.3552 0 0.0961 ...
## ..- attr(*, "header")= chr "Coefficients"
## ..- attr(*, "formats")= chr [1:3] "%s" "%5f" "%5f"
## ..- attr(*, "description")= chr "glm coefficients"
## $ random_coefficients_table : NULL
## $ coefficients_table_multinomials_with_class_names: NULL
## $ standardized_coefficient_magnitudes :Classes 'H2OTable' and 'data.frame': 36 obs. of 3 variables:
## ..$ names : chr [1:36] "GBM_2_AutoML_20210725_011103" "GBM_grid__1_AutoML_20210725_011103_model_18" "GBM_grid__1_AutoML_20210725_011103_model_9" "GBM_grid__1_AutoML_20210725_011103_model_19" ...
## ..$ coefficients: num [1:36] 0.701 0.657 0.634 0.437 0.405 ...
## ..$ sign : chr [1:36] "POS" "POS" "POS" "POS" ...
## ..- attr(*, "header")= chr "Standardized Coefficient Magnitudes"
## ..- attr(*, "formats")= chr [1:3] "%s" "%5f" "%s"
## ..- attr(*, "description")= chr "standardized coefficient magnitudes"
## $ variable_importances :Classes 'H2OTable' and 'data.frame': 36 obs. of 4 variables:
## ..$ variable : chr [1:36] "GBM_2_AutoML_20210725_011103" "GBM_grid__1_AutoML_20210725_011103_model_18" "GBM_grid__1_AutoML_20210725_011103_model_9" "GBM_grid__1_AutoML_20210725_011103_model_19" ...
## ..$ relative_importance: num [1:36] 0.701 0.657 0.634 0.437 0.405 ...
## ..$ scaled_importance : num [1:36] 1 0.936 0.903 0.624 0.577 ...
## ..$ percentage : num [1:36] 0.162 0.1516 0.1463 0.101 0.0935 ...
## ..- attr(*, "header")= chr "Variable Importances"
## ..- attr(*, "formats")= chr [1:4] "%s" "%5f" "%5f" "%5f"
## ..- attr(*, "description")= chr ""
## $ lambda_best : num 0.00311
## $ alpha_best : num 0.5
## $ best_submodel_index : int 0
## $ lambda_1se : num 0.00311
## $ lambda_min : num -1
## $ lambda_max : num -1
## $ dispersion : num 0
## $ coefficients : Named num [1:37] 0.3863 0.183 0.0834 0 0.0228 ...
## ..- attr(*, "names")= chr [1:37] "Intercept" "GBM_2_AutoML_20210725_011103" "GBM_grid__1_AutoML_20210725_011103_model_2" "GBM_4_AutoML_20210725_011103" ...